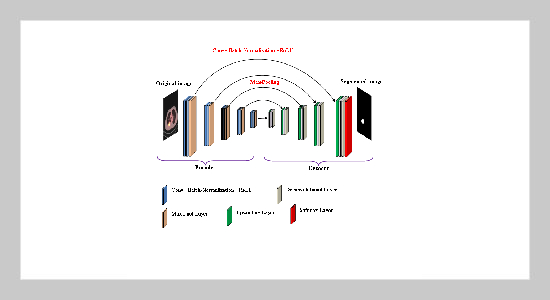
- [1] B. Le Goff, J.-M. Berthelot, and Y. Maugars, (2015) “Échographie du thorax antérieur [Ultrasound of the anterior thorax]" Revue du Rhumatisme Monographies 82(2): 83–87. DOI: 10.1016/j.monrhu.2015.02.005.
- [2] C. Jani, D. C. Marshall, H. Singh, R. Goodall, J. Shalhoub, O. Al Omari, and C. C. Thomson, (2021) “Lung cancer mortality in Europe and the USA between 2000 and 2017: An observational analysis" ERJ Open Re search 7(4): DOI: 10.1183/23120541.00311-2021.
- [3] P. B. Bach, J. R. Jett, U. Pastorino, M. S. Tockman, S. J. Swensen, and C. B. Begg, (2007) “Computed tomogra phy screening and lung cancer outcomes" JAMA 297(9): 953–961. DOI: 10.1001/jama.297.9.953.
- [4] E. B. Jemia, H. Kamoun, S. Louhaichi, H. Smadhi, D. Greb, I. Akrout, and M. L. Megdiche, (2017) “Variation du profilhistologique du cancer du poumon durant les 25 dernières années dans un service depneumologie à Tunis [Variation of the histological profile of lung cancer during the last 25 years in a pulmonology department in unis]" Revue des Maladies Respiratoires 34: A79. DOI: 10.1016/j.rmr.2016.10.168.
- [5] Y. Men, Z. Hui, J. Liang, Q. Feng, D. Chen, H. Zhang, and L. Wang, (2016) “Further understanding of an un common disease of combined small cell lung cancer: Clinical features and prognostic factors of 114 cases" Chinese Journal of Cancer Research 28(5): 486. DOI: 10.21147/j.issn.1000-9604.2016.05.03.
- [6] B. Padovani, J. Mouroux, L. Seksik, S. Chanalet, J. Sedat, C. Rotomondo, and J. J. Serres, (1993) “Chest wall invasion by bronchogenic carcinoma: Evaluation with MRimaging" Radiology 187(1): 33–38. DOI: 10.1148/ radiology.187.1.8451432.
- [7] N.Suzuki, T. Saitoh, and S. Kitamura, (1993) “Tumor invasion of the chest wall in lung cancer: Diagnosis with US"Radiology187(1): 39–42. DOI: 10.1148/radiology.187.1.8451433.
- [8] S. J. Swensen, R. W. Viggiano, D. E. Midthun, N. L. Müller, A. Sherrick, K. Yamashita, and A. L. Weaver, (2000) “Lung nodule enhancement at CT: Multicen ter study" Radiology 214(1): 73–80. DOI: 10.1148/radiology.214.1.r00ja1473.
- [9] D. F. Yankelevitz, A. P. Reeves, W. J. Kostis, B. Zhao, and C. I. Henschke, (2000) “Small pulmonary nodules: Volumetrically determined growth rates based on CT evaluation" Radiology 217(1): 251–256. DOI: 10.1148/radiology.217.1.r00oc33251.
- [10] G. Ferretti, A. Jankowski, A. Calizzano, D. Moro Sibilot, and J. P. Vuillez, (2008) “Imagerie radiologique et TEP Scanner dans les cancers du poumon [Radiological imaging and PET scans in lung cancer]" Journal de Radiologie 89(3): 387–402. DOI: 10.1016/S0221-0363(08)89016-6.
- [11] N. A. Dewan, S. D. Reeb, N. C. Gupta, L. S. Go bar, and W. J. Scott, (1995) “PET-FDG imaging and transthoracic needle lung aspiration biopsy in evaluation of pulmonary lesions: A comparative risk-benefit analysis" Chest 108(2): 441–446. DOI: 10.1378/chest.108.2.441.
- [12] J. E. Bibault, A. Oudoux, J. Durand-Labrunie, X. Mirabel, É. Lartigau, and H. Kolesnikov-Gauthier, (2015) “TEP et radiothérapie stéréotaxique pulmonaire : rôles dans la préparation du traitement et le suivi de la mal adie [Positron emission tomography and stereotactic body radiation therapy for lung cancer: From treatment planning to response evaluation]" Cancer/Radiothérapie 19(8): 790–794. DOI: 10.1016/j.canrad.2015.05.027.
- [13] S. S. Gambhir, J. Czernin, J. Schwimmer, D. H. Sil verman, R. E. Coleman, and M. E. Phelps, (2001) “A tabulated summary of the FDG PET literature" Journal of Nuclear Medicine 42(5 suppl): 1S–93S.
- [14] M. K. Gould, C. C. Maclean, W. G. Kuschner, C. E. Rydzak,andD.K.Owens,(2001)“Accuracyofpositron emission tomography for diagnosis of pulmonary nodules and mass lesions: A meta-analysis" JAMA 285(7): 914 924. DOI: 10.1001/jama.285.7.914.
- [15] C. Perrotin, P. Lemeunier, D. Grahek, T. Molina, A. Petino, M. Alifano, F. Bellenot, P. Magdeleinat, J. N. Talbot, and J. F. Regnard, (2005) “Résultats de la TEP [18F]-FDG dansla stadification préopératoire des tumeurs pulmonaires [Results of FDG-PET scanning in the pre operative staging of broncho-pulmonary tumors]" Revue des Maladies Respiratoires 22(4): 579–585. DOI: 10.1016/s0761-8425(05)85610-4.
- [16] J. F. Vansteenkiste, S. G. Stroobants, P. J. Dupont, P. R. De Leyn, E. K. Verbeken, G. J. Deneffe, and M. G. Demedts, (1999) “Prognostic importance of the standardized uptake value on 18F-fluoro-2-deoxy-glucose–positron emission tomography scan in non–small-cell lung cancer: An analysis of 125 cases" Journal of Clinical Oncology 17(10): 3201–3206.
- [17] M.Vallières, C. R. Freeman, S. R. Skamene, and I. El Naqa, (2015) “A radiomics model from joint FDG-PET and MRI texture features for the prediction of lungmetas tases in soft-tissue sarcomas of the extremities" Physics in Medicine & Biology 60(14): 5471. DOI: 10.1088/0031-9155/60/14/5471.
- [18] Z. Li, J. Zhang, T. Tan, X. Teng, X. Sun, H. Zhao, and G. Litjens, (2020) “Deep learning methods for lung cancer segmentation in whole-slide histopathology images—the acdc@ lunghp challenge 2019" IEEE Journal of Biomedical and Health Informatics 25(2): 429 440. DOI: 10.1109/JBHI.2020.3039741.
- [19] H. Hu, Q. Li, Y. Zhao, and Y. Zhang, (2020) “Parallel deep learning algorithms with hybrid attention mechanism for image segmentation of lung tumors" IEEE Transactions on Industrial Informatics 17(4): 2880 2889. DOI: 10.1109/TII.2020.3022912.
- [20] P. Li, S. Wang, T. Li, J. Lu, Y. HuangFu, and D. Wang. A large-scale CT and PET/CT dataset for lung cancer diagnosis (Lung-PET-CT-Dx) [Data set]. 2020. DOI: 10.7937/TCIA.2020.NNC2-0461.
- [21] M. Wang, X. Zhou, M. Jin, Y. Zhang, L. Liu, and G. Huang, (2024) “Multiroimix: A data augmentation method for PET/CT multimodal medical images" Journal of Medical and Biological Engineering 44(3): 366 374. DOI: 10.1007/s40846-024-00862-y.
- [22] A. Klaengkaew, S. Sutthigran, N. Thammasiri, K. Yuwatanakorn, C. Thanaboonnipat, S. Ponglowha pan, and N. Choisunirachon, (2021) “The evaluation of non-anesthetic computed tomography for detection of pulmonary parenchyma in feline mammary gland carcinoma: a preliminary study" BMC Veterinary Research 17(1): 237. DOI: 10.1186/s12917-021-02950-6.
- [23] X. Zhang, G. Xu, X. Wu, W. Liao, L. Xiao, Y. Jiang, and H. Xing, (2024) “Fast-SegNet: Fast semantic seg mentation network for small objects" Multimedia Tools and Applications 83(34): 81039–81055. DOI: 10.1007/s11042-024-18829-1.
- [24] R. Pang, H. Tan, Y. Yang, X. Xu, N. Liu, and P. Zhang. “A Novel SegNet Model for Crack Image Semantic Segmentation in Bridge Inspection”. In: Advances in Knowledge Discovery and Data Mining. PAKDD 2024. Ed. by D. Yang, X. Xie, V. Tseng, J. Pei, J. Huang, and J. Lin. 14647. Lecture Notes in Computer Science. Singapore: Springer, 2024, 26. DOI: 10.1007/978-981-97-2259-4_26.
- [25] N. ¸Sahin, N. Alpaslan, andD.Hanbay,(2022)“Robust optimization of SegNet hyperparameters for skin lesion segmentation" Multimedia Tools and Applications 81(25): 36031–36051. DOI: 10.1007/s11042-021-11032-6.
- [26] Y. Zhang, (2025) “Image denoising based on deep feature fusion and U-Netnetwork "Journal of Applied Science and Engineering 28(10): 2077–2085. DOI: 10.6180/jase.202510_28(10).0020.
- [27] H.ChenandZ.Xu,(2021) “A new end-to-end network model for medical image segmentation" Journal of Ap plied Science and Engineering 24(2): 207–213. DOI: 10.6180/jase.202104_24(2).0009.
- [28] G. Athanasiou, J. L. Arcos, and J. Cerquides, (2023) “Enhancing medical image segmentation: Ground truth optimization through evaluating uncertainty in expert annotations" Mathematics 11(17): 3771. DOI: 10.3390/math11173771.
- [29] S. Alqazzaz, X. Sun, X. Yang, and L. Nokes, (2019) “Automatedbraintumorsegmentationonmulti-modalMR image using SegNet" Computational Visual Media 5(2): 209–219. DOI: 10.1007/s41095-019-0139-y.
- [30] G. Di Leo and F. Sardanelli, (2020) “Statistical significance: p value, 0.05 threshold, and applications to radiomics—reasons for a conservative approach" Euro pean Radiology Experimental 4(1): 18. DOI: 10.1186/s41747-020-0145-y.
- [31] J. M. Rogasch, N. Frost, S. Bluemel, L. Michaels, T. Penzkofer, M. von Laffert, and C. Furth, (2021) “FDG-PET/CT for pretherapeutic lymph node staging in non-small cell lung cancer: a tailored approach to the ESTS/ESMO guideline workflow" Lung Cancer 157: 66–74. DOI: 10.1016/j.lungcan.2021.05.003.
- [32] S. N. Kumar, P. M. Bruntha, S. I. Daniel, J. A. Kirubakar, R. E. Kiruba, S. Sam, and S. I. A. Pandian. “Lung nodule segmentation using U-Net”. In: 2021 7th International Conference on Advanced Computing and Communication Systems (ICACCS). 1. 2021, 420 424. DOI: 10.1109/ICACCS51430.2021.9441977.
- [33] G.M.Alshmrani,Q.Ni,R.Jiang,andN.Muhammed, (2023) “Hyper-dense_lung_seg: Multimodal-fusion-based modified U-Net for lung tumor segmentation using multimodality of CT-PET scans" Diagnostics 13(22): 3481. DOI: 10.3390/diagnostics13223481.
- [34] J. Park, S. K. Kang, D. Hwang, H. Choi, S. Ha, J. M. Seo, and J. S. Lee, (2023) “Automatic lung cancer seg mentation in [18F] FDG PET/CT using a two-stage deep learning approach" Nuclear Medicine and Molecular Imaging 57(2): 86–93. DOI: 10.1007/s13139-022 00745-7.
- [35] L. Zhou, C. Wu, Y. Chen, and Z. Zhang, (2024) “Multitask connected U-Net: automatic lung cancer segmentation from CT images using PET knowledge guidance" Frontiers in Artificial Intelligence 7: 1423535. DOI: 10.3389/frai.2024.1423535.
- [36] G. Saimassay, M. Begenov, U. Sadyk, R. Baimuka shev, A. Maratov, and B. Omarov, (2024) “Enhanced U-Net architecture for lung segmentation on computed tomography and X-ray images" International Journal of Advanced Computer Science and Applications 15(5): DOI: 10.14569/IJACSA.2024.0150594.
- [37] A. Ben Slama, Y. Amri, A. Fnaiech, and H. Sahli, (2025) “Automated ECG arrhythmia classification using hybrid CNN-SVM architectures" Journal of Electronic Science and Technology 23(3): 100316. DOI: 10.1016/j.jnlest.2025.100316.
- [38] Y. Amri, A. Ben Slama, Z. Mbarki, R. Selmi, and H. Trabelsi, (2025) “Automatic glioma segmentation based on efficient U-Net model using MRI images" Intelligence-Based Medicine: 100216. DOI: 10.1016/j.ibmed.2025.100216.
- [39] A. Ben Slama, Y. Amri, S. Barbaria, H. B. Rahmouni, and H. Trabelsi, (2025) “Lung diseases classification using pre-trained based deep learning model and support vector machine" Polish Journal of Medical Physics andEngineering31(3): 178–194. DOI: 10.2478/pjmpe-2025-0021.
- [40] I. Domingues, G. Pereira, P. Martins, H. Duarte, J. Santos, and P. H. Abreu, (2020) “Using deep learning techniques in medical imaging: a systematic review of applications on CT and PET" Artificial Intelligence Review 53(6): 4093–4160. DOI: 10.1007/s10462-019 09850-2.
- [41] S. Yin, H. Li, L. Teng, and et al., (2024) “Brain CT image classification based on mask R-CNN and attention mechanism" Scientific Reports 14: 29300. DOI: 10.1038/s41598-024-78566-1.
- [42] S. Yin, L. Wang, T. Chen, H. Huang, J. Gao, J. Zhang, M. Liu, P. Li, and C. Xu, (2025) “LKAFormer: A lightweight Kolmogorov-Arnold transformer model for image semantic segmentation" ACM Transactions on Intelligent Systems and Technology:
- [43] Q. Shi, S. Yin, K. Wang, L. Teng, and H. Li, (2021) “Multichannel convolutional neural network-based fuzzy active contour model for medical image segmentation" Evolving Systems 13: 535–549. DOI: 10.1007/s12530 021-09392-3.